At Cell Surface Bio, we validate that our MAbs work, so you don’t have to. CSB’s VeRSaMAbs are recombinant monoclonal antibodies that are designed, produced, and validated in-house with the highest quality control standards.1-3 Our extensive validation and specificity profiling powers the CSB VeRSaMAb catalog with antibodies that are validated to bind your desired target. You can expect a dependable, reproducible product from CSB every time.
Validated Applications
Antibodies are critical in evaluating the relative expression, localization, and interaction of targets when used in immunoassays such as flow cytometry, immunofluorescence, western blots, ELISA, immunohistochemistry/cytochemistry, and more. Each of these applications involves different assay conditions and target conformations, so the ability of a specific MAb to work in one application does not guarantee that it will work in another. In fact, the ability of a monoclonal antibody to work in one application (e.g., western blot of denatured protein) sometimes precludes the ability of the same one to work in another application (e.g., flow cytometry of a conformational epitope on the cell surface). The epitope that an antibody binds to can be altered by experimental conditions, which is why at CSB we test each MAb to determine which applications and experimental conditions actually work.
As part of our standard antibody validation process, CSB currently conducts the following validation experiments for each of our antibodies:
- Flow cytometry extracellular (unfixed)
- Flow cytometry intracellular (fixed with paraformaldehyde and saponin)
- Immunofluorescence extracellular (fixed with paraformaldehyde)
- Immunofluorescence intracellular (fixed with paraformaldehyde and Triton X-100)
- (more applications coming soon!)
Click here for our standardized protocols.
Flow Cytometry (FC)
Flow cytometry is used to measure the expression of cellular markers. Analysis of protein expression is commonly conducted by measuring the fluorescence intensity of single cells in suspension, following incubation with an antibody against the target of interest. Fluorescent labeling of antibodies can be achieved either through direct fluorophore conjugation to the primary antibody or from fluorescently conjugated secondary antibodies.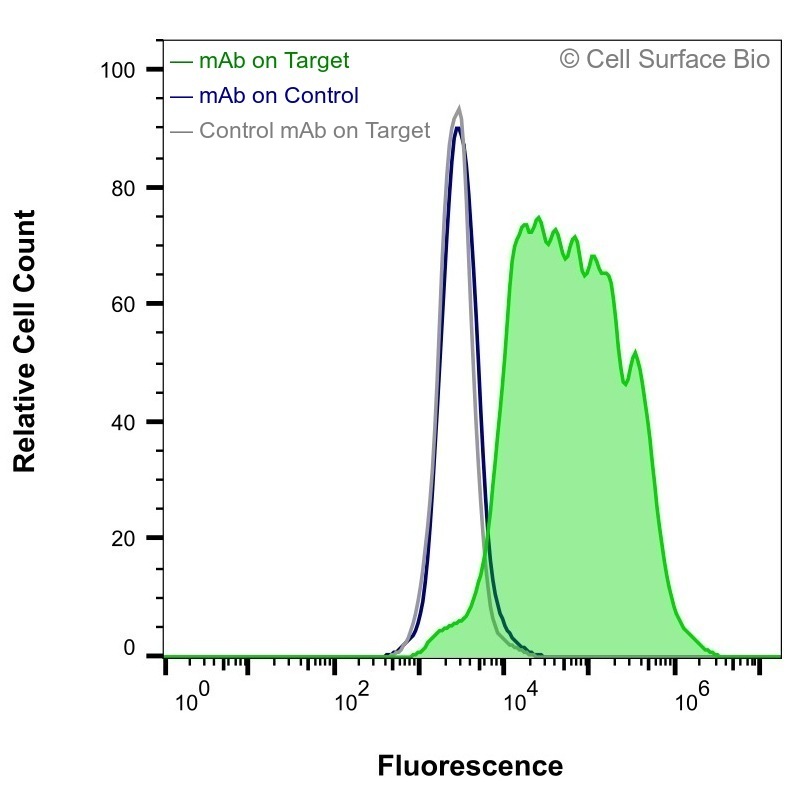
Figure 1. HEK-293F cells transiently transfected with human CD7 were stained with CD7 Antibody (CSB0011) (green) or isotype control antibody (gray), followed by AlexaFluor 647-conjugated anti-mouse IgG secondary antibody. HEK-293F cells transiently transfected with an empty control vector were also stained with CD7 Antibody (CSB0011) (blue).
Immunofluorescence (IF)
Immunofluorescence is used to visualize the localization of a target within cells. Cultured cells are commonly fixed with paraformaldehyde, incubated with a primary antibody, incubated with a secondary antibody conjugated to a fluorophore (indirect immunofluorescence), and then visualized via microscopy. IF can provide insight into the relative expression, subcellular localization, and cell-type specific expression of target antigens.
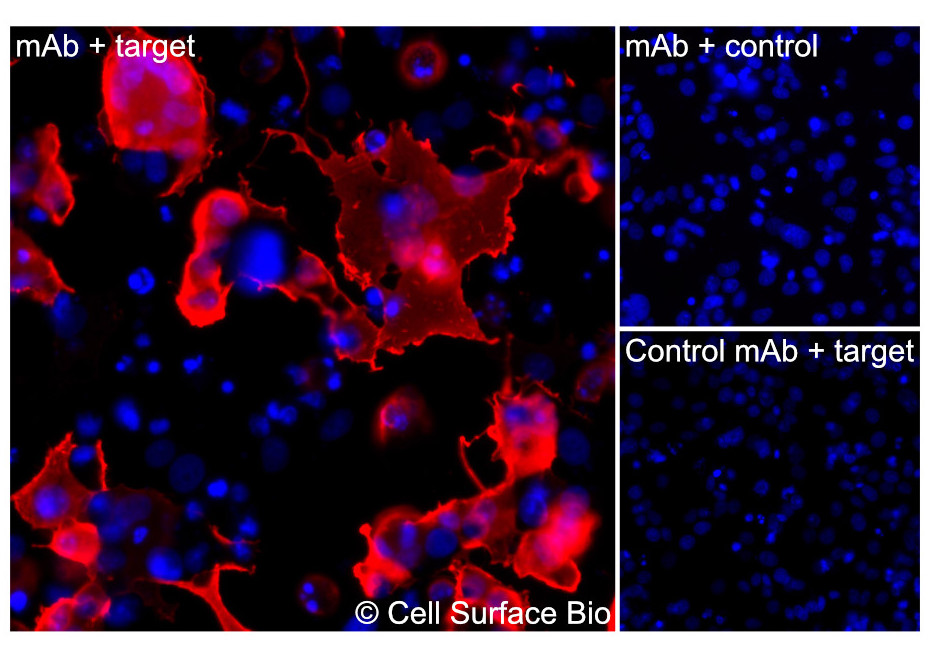
Figure 2. (A) COS-7 cells transiently transfected with human CD7 were stained with CD7 Antibody (CSB0011) followed by AlexaFluor 647 anti-mouse IgG secondary antibody (red) and DAPI (blue). (B) COS-7 cells transiently transfected with an empty control vector stained with CD7 Antibody. (C) Isotype control: COS-7 cells transfected with human and stained with control MAb.
CSB’s Antibody Validation Experiments
2 cell lines & 2 controls each
CSB’s VeRSaMAbs are validated by analyzing antibody binding in cell lines overexpressing the target of interest. All CSB antibodies are tested in a minimum of two cell lines (e.g. HEK-293F and COS-7), which enables us to differentiate antibody binding due to endogenous expression from any non-specific binding (Figure 4). We transfect cell lines with plasmid DNA vector encoding the target of interest and run an empty DNA vector transfection simultaneously as a negative control. We also stain cells with an isotype-matched control MAb. Every assay therefore evaluates the following conditions:
- Antibody signal in target-transfected cells
- Antibody signal in control-transfected cells (negative control)
- Isotype control MAb signal in target-transfected cells (negative control)
|
Figure 3. Flow cytometry C5AR antibody (CSB0027) transiently transfected HEK-293F cells. Green = MAb on target, Blue= MAb on Control, Gray= Control MAb on Target |
Figure 4. Flow cytometry EGFR antibody (CSB0008) EGFR endogenously expressed in HEK-293F cells. Green = MAb on target, Blue= MAb on Control, Gray= Control MAb on target, also tested in COS7 cells |
Extracellular vs. Intracellular Epitope Localization
Localization of the epitope (where the antibody binds) impacts detection of the target and the conditions required for detection. For example, if an antibody binds to an intracellular epitope, then the antibody cannot bind without cell membrane permeabilization (Figure 5).
%20Live%20extracellular%20flow%20cytometry.jpg) |
%20Fixed%20(4%25%20PFA)%20and%20Permeabilized%20(0.02%25%20Saponin)%20Intracellular%20flow%20cytometry.jpg) |
|
Figure 5. SLC4A1 antibody (CSB0146) Live extracellular flow cytometry (left), SLC4A1 antibody (CSB0146) Fixed (4% PFA) and Permeabilized (0.02% Saponin) Intracellular flow cytometry (right). |
|
To identify the subcellular localization of the antigen’s epitope, every CSB antibody is tested under both extracellular and intracellular binding conditions in both flow cytometry and immunofluorescence assays. Extracellular binding evaluates cell surface expression using non-permeabilized conditions that keep the cell membrane intact. Intracellular binding conditions are evaluated by permeabilizing cells with detergents, allowing antibodies to bind to epitopes on the inside of the cell.
Fixed vs Unfixed Binding Conditions
Fixation using aldehyde-based solutions is often used to cross-link proteins to prevent the loss of antigens. Fixation using formalin-based solutions is often used to preserve tissue and cell morphology. However, fixation can also lead to conformational changes or the masking of epitopes, preventing proper antibody binding. CSB tests all antibodies under unfixed conditions, fixed conditions, and permeabilized conditions (using two different detergents) for validation of each application
- Flow cytometry:
- Extracellular staining: Unfixed conditions (live cells to detect the native epitope)
- Intracellular staining: Fixed conditions (paraformaldehyde and saponin)
- Immunofluorescence:
- Extracellular staining: Fixed conditions (paraformaldehyde)
- Intracellular staining: Fixed conditions (paraformaldehyde and Triton X-100)
At Cell Surface Bio, we want every experiment you run with our reagents to work.
Click here for our standardized protocols.
References
(1) Taussig, M. J.; Fonseca, C.; Trimmer, J. S. Antibody Validation: A View from the Mountains. N Biotechnol 2018, 45, 1–8. https://doi.org/10.1016/j.nbt.2018.08.002.
(2) Dove, A. Agreeable Antibodies: Antibody Validation Challenges and Solutions. Science 2017. https://doi.org/10.1126/science.opms.p1700116.
(3) Weller, M. G. Ten Basic Rules of Antibody Validation. Anal ChemInsights 2018, 13, 117739011875746. https://doi.org/10.1177/1177390118757462.
(4) Baker, M. Antibodies Are the Workhorses of Biological Experiments, but They Are Littering the Field with False Findings. A Few Evangelists Are Pushing for Change. 3.
(5) Bordeaux, J.; Welsh, A. W.; Agarwal, S.; Killiam, E.; Baquero, M. T.; Hanna, J. A.; Anagnostou, V. K.; Rimm, D. L. Antibody Validation. BioTechniques 2010, 48 (3), 197–209. https://doi.org/10.2144/000113382.
(6) Voskuil, J. L. A. The Challenges with the Validation of Research Antibodies. F1000Res 2017, 6, 161. https://doi.org/10.12688/f1000research.10851.1.
(7) Voskuil, J. Commercial Antibodies and Their Validation [Version 2; Peer Review: 3 Approved]. F1000Research 2014, 3 (232). https://doi.org/10.12688/f1000research.4966.2.
(8) Weller, M. G. Quality Issues of Research Antibodies. Anal ChemInsights 2016, 11, ACI.S31614. https://doi.org/10.4137/ACI.S31614.
(9) Bradbury, A.; Plückthun, A. Reproducibility: Standardize Antibodies Used in Research. Nature 2015, 518 (7537), 27–29. https://doi.org/10.1038/518027a.

%20transiently%20transfected%20HEK-293F%20cells.%20Green%20=%20mAb%20on%20target.jpg)
%20Endogenously%20expressed%20in%20HEk%20293F%20cells..jpg)